Flipons help Intrinsically Disordered Proteins Fold into Cellular Machines
Different DNA Structures Nucleate the Folding of Proteins to form specific complexes that modulate the readout of genetic information
Self-organization is inherent in the way Nature has engineered cells. The folding of proteins induced by different DNA structures encoded by sequences called flipons is just one example.”
CHARLESTOWN, MA, UNITED STATES, February 2, 2026 /EINPresswire.com/ -- In a paper published today, researchers at InsideOutBio have used AI to examine how cells fold proteins into active complexes, using alternative DNA structures to seed cellular machines. The paper entitled “The Chromaverse Is Colored by Triplexes Formed Through the Interactions of Noncoding RNAs with HNPRNPU, TP53, AGO, REL Proteins, Intrinsically-Disordered Regions, and Flipons” by Alan Herbert focuses on three standard triplexes made by wrapping an RNA around a DNA duplex. The triplexes help fold proteins in a way that dynamically organizes DNA within a cell, resulting in a multitude of chromatin states that collectively constitute the chromaverse of possibilities.— Alan Herbert
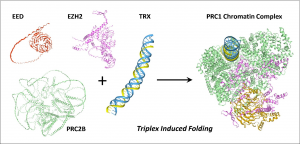
Using AlphaFold Version 3, the paper reveals how disordered proteins can recognize the triplex to form organized structures that modulate the readout of genetic information. The assemblies are formed dynamically, in contrast to the well-known lock-and-key mechanism based on B-DNA, where proteins bind with high affinity to specific bases in the two-stranded Watson-Crick duplex. In contrast, the three-stranded RNA/DNA structures induce folding of disordered proteins. The bound proteins then initiate folding and binding of other proteins to set in motion the assembly of cellular machines, each with specific functions. The outcome depends on what proteins are available in the cell at that moment. It also varies with the extent to which these proteins are modified by other cellular processes. Some modifications promote binding, whereas others might prevent interaction. In some cases, modifications alter how a disordered region folds, thereby favoring the formation of a complex that performs one function over another with different properties.
Specific examples of how triplexes direct folding are given in the paper. These case studies include proteins implicated in cancer, such as p53, and proteins involved in inflammatory responses, such as the REL family. Additionally, it was possible to demonstrate the assembly of multi-component complexes involved in chromatin modification, such as polycomb repressive complexes 1 and 2 (PRC1 and PRC2). A description of experimentally confirmed triplex interactions with various proteins is also presented in the paper. The value of this approach is that highly detailed structures are developed that can be viewed by anyone with a freely available structure viewer, enabling independent assessment of interactions described and providing a framework to guide experimentation.
This use of alternative structures encoded by sequences called flipons likely dates back to the early origins of life, when different DNA and RNA folds enabled a variety of chemistries that facilitated the replication of these elements. In the modern world, this ancient readout of genetic information, using flipon structures to guide protein folding, and base-specific binding to DNA sequences work together to produce specialized cells that can dynamically respond to challenges that threaten an organism’s survival. The paper provides examples of ways in which these strategies combine to protect hosts against invasive replicants, such as viruses.
The paper captures recent changes in our understanding of how cells work. We are moving beyond models based solely on the right-handed B-DNA conformation described by Watson and Crick. Instead, current research focuses on how dynamical changes in DNA and RNA structures affect cell biology and disease processes. These advances have been enabled by substantial investment in the Human Genome Project, by technologies that enable analysis of the large datasets produced, and by recent advances in AI. Progress is exemplified by a new appreciation of the role of the genome's repetitive elements. These elements were once considered non-informative because, as B-DNA, each repeat is so frequent. However, flipons are only encoded by repeat sequences. By changing their structure, flipons can switch on different pathways. This is exemplified by cellular Z-DNA and Z-RNAs, which regulate immune responses to viruses.
InsideOutBio is a start-up focused on developing a novel class of proprietary therapeutics to ‘light up tumors for the immune system. These statements about InsideOutBio comply with Safe-Harbor laws. They are forward-looking and involve known and unknown risks and uncertainties. They are not guarantees of future performance, and undue reliance should not be placed on them.
Alan Herbert
InsideOutBio, Inc
+1 617-584-0360
email us here
Visit us on social media:
LinkedIn
X
Legal Disclaimer:
EIN Presswire provides this news content "as is" without warranty of any kind. We do not accept any responsibility or liability for the accuracy, content, images, videos, licenses, completeness, legality, or reliability of the information contained in this article. If you have any complaints or copyright issues related to this article, kindly contact the author above.